Researchers at Colorado State University have developed a new imaging technique that shows key cellular interactions over time and space in a way that was not previously possible. The work could one day help in efforts to control “gene expression” in medicine, such as turning off genes that cause cancer.
Described in Science Advances, the research was performed with a new microscopy approach that captures interactions in the cell between DNA and proteins over time, similar to movies. Previous techniques to observe these interactions were only able to capture single-image snapshots, providing just a limited glimpse into key chemical modifications and cellular machinery.
Associate Professor Tim Stasevich led the research from the Department of Biochemistry and Molecular Biology on campus. He said the ability to capture a sequence of events as movies instead of stills offers a new, powerful way to observe the impact chemical modifications have on proteins as they interact with DNA to accomplish the tasks needed for life.
DNA contains the information needed to build and maintain organisms – it is the genetic blueprint that controls how cells function, grow and divide. However, the ways living cells regulate and use those blueprints is a continuing question for researchers.
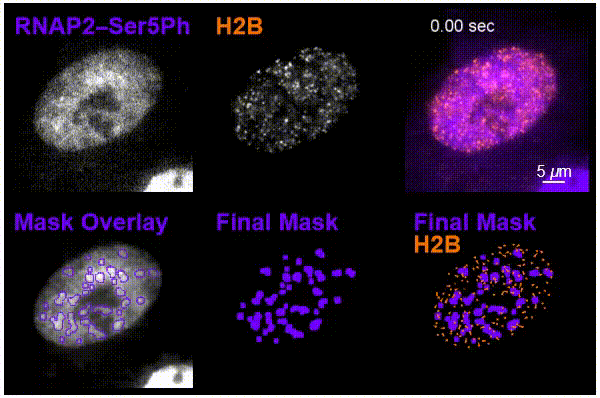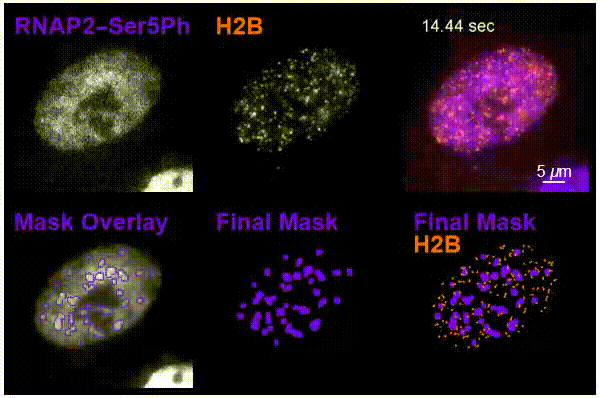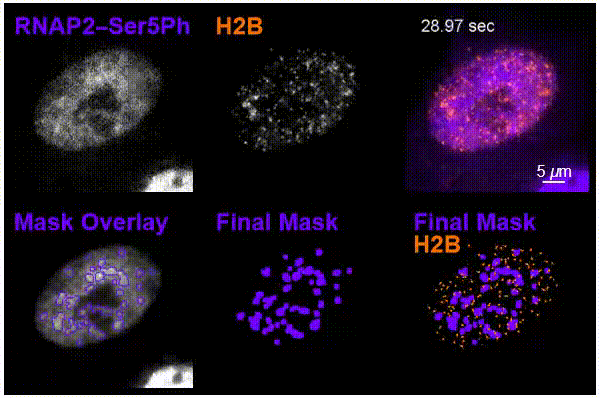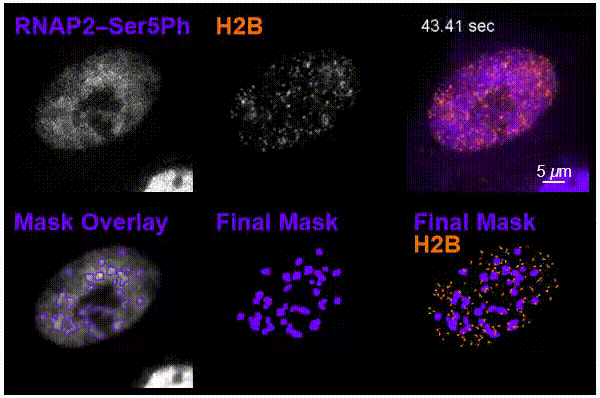
The team was particularly interested in tracking interactions in cells around nucleosomes – a complex of DNA wrapped around a core of histone proteins – and RNA pol II, which transcribes DNA into the mRNA needed to create proteins.
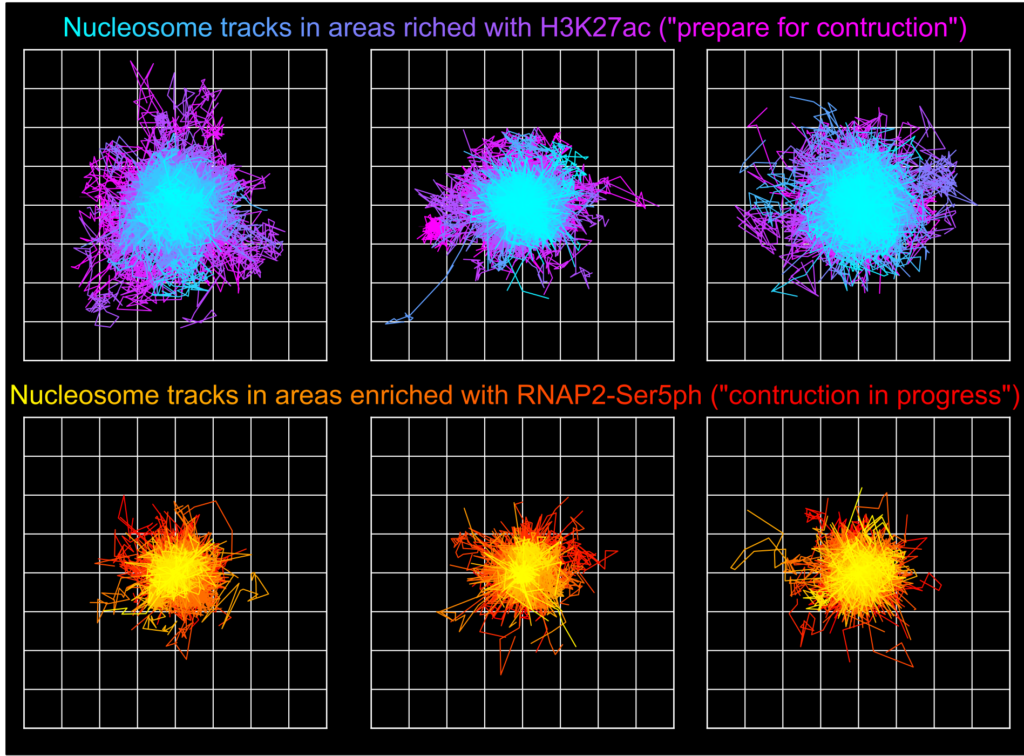
Using the new imaging approach, the team was able to track nucleosomes down to the single-molecule level in a living cell while simultaneously monitoring local chemical modifications to both nucleosomes and RNA pol II. This was all done in real time and in a way that was not possible before. The results of the research showed that the dynamics of nucleosomes were different depending on the specific nearby chemical modifications, meaning the modifications are not typically enriched together inside living cells, Stasevich said.
“Our work suggests cells have specialized areas with distinct functions and dynamics that are separated from one another in space and/or time – one for preparing DNA for reading and another for the construction of mRNA from DNA,” he said. “We were now able to better see these as – for example – signs at a building site that read ‘prepare for construction’ or ‘construction in progress,’ depending on the situation.”
Stasevich added that future work would explore how nucleosomes could be modified with different reactions and in distinct locations in the cell.
“I envision many future experiments where we look at the different versions of these modifications across several instances to see how they can be used and combined to fine-tune nucleosome mobility and gene expression,” he said.
Matthew Saxton is the first author on the paper and recently graduated with his Ph.D. in biochemistry after studying with Stasevich. He said this interdisciplinary research was difficult to pull off.
“To do this, cells had to be genetically manipulated and then imaged on a complex microscope. And we then had to write code to then analyze a large volume of data,” he said. “It was a team effort across the biochemistry, biophysics and microscopy disciplines.”
He added that this research provides insight into the guiding principles and tenets behind how life functions with broad future applications.
“We wanted to observe how genetic material responds when a cell activates transcription or marks areas for future transcription. This gives us much better information when we attempt to correct aberrant and disease-causing behavior,” he said.